Dimension reduction and heatmaps
Dimension reduction
We start loading the tissue gene expression dataset:
# library(devtools) install_github('dagdata','genomicsclass')
library(dagdata)
data(tissuesGeneExpression)
library(Biobase)
## Loading required package: BiocGenerics
## Loading required package: methods
## Loading required package: parallel
##
## Attaching package: 'BiocGenerics'
##
## The following objects are masked from 'package:parallel':
##
## clusterApply, clusterApplyLB, clusterCall, clusterEvalQ,
## clusterExport, clusterMap, parApply, parCapply, parLapply,
## parLapplyLB, parRapply, parSapply, parSapplyLB
##
## The following object is masked from 'package:stats':
##
## xtabs
##
## The following objects are masked from 'package:base':
##
## anyDuplicated, append, as.data.frame, as.vector, cbind,
## colnames, do.call, duplicated, eval, evalq, Filter, Find, get,
## intersect, is.unsorted, lapply, Map, mapply, match, mget,
## order, paste, pmax, pmax.int, pmin, pmin.int, Position, rank,
## rbind, Reduce, rep.int, rownames, sapply, setdiff, sort,
## table, tapply, union, unique, unlist
##
## Welcome to Bioconductor
##
## Vignettes contain introductory material; view with
## 'browseVignettes()'. To cite Bioconductor, see
## 'citation("Biobase")', and for packages 'citation("pkgname")'.
rownames(tab) <- tab$filename
t <- ExpressionSet(e, AnnotatedDataFrame(tab))
t$Tissue <- factor(t$Tissue)
colnames(t) <- paste0(t$Tissue, seq_len(ncol(t)))
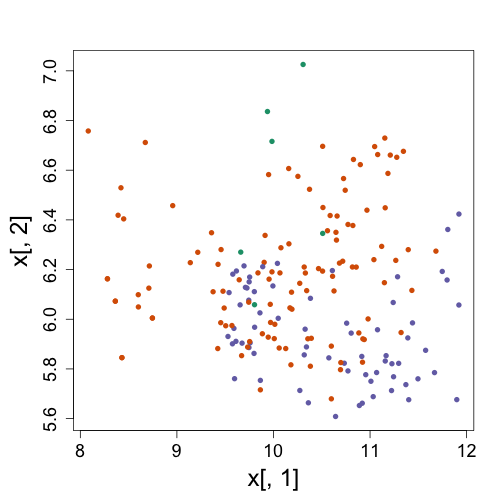
As we noticed in the end of the clustering section, we weren’t able to see why the k-means algorithm defined a certain set of clusters using only the first two genes.
x <- t(exprs(t))
km <- kmeans(x, centers = 3)
library(rafalib)
## Loading required package: RColorBrewer
mypar()
plot(x[, 1], x[, 2], col = km$cluster, pch = 16)
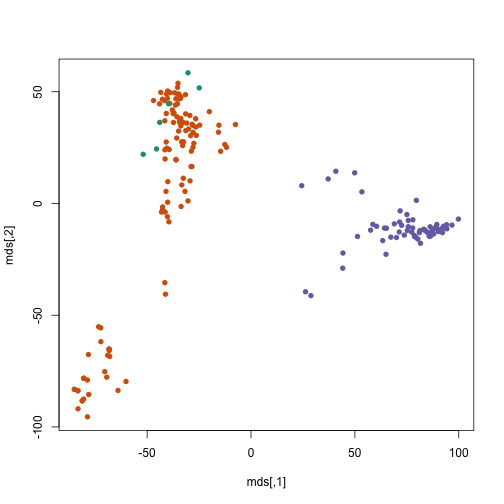
Instead of the first two genes, let’s use the multi-dimensional
scaling algorithm which Rafa introduced in the lectures. This is a
projection from the space of all genes to a two dimensional space,
which mostly preserves the inter-sample distances. The cmdscale
function in R takes a distance object and returns a matrix which has
two dimensions (columns) for each sample.
mds <- cmdscale(dist(x))
plot(mds, col = km$cluster, pch = 16)
We can also plot the names of the tissues with the color of the cluster.
plot(mds, type = "n")
text(mds, colnames(t), col = km$cluster)
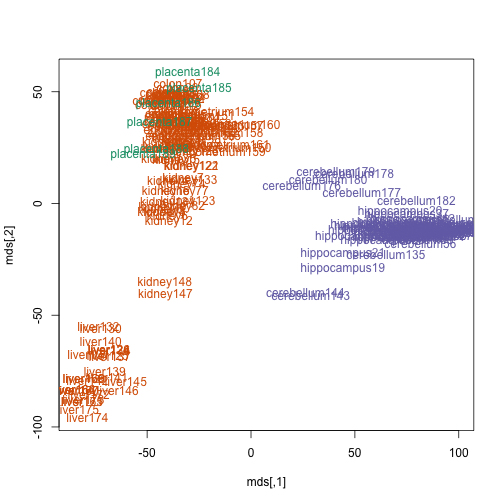
…or the names of the tissues with the color of the tissue.
plot(mds, type = "n")
text(mds, colnames(t), col = as.fumeric(t$Tissue))

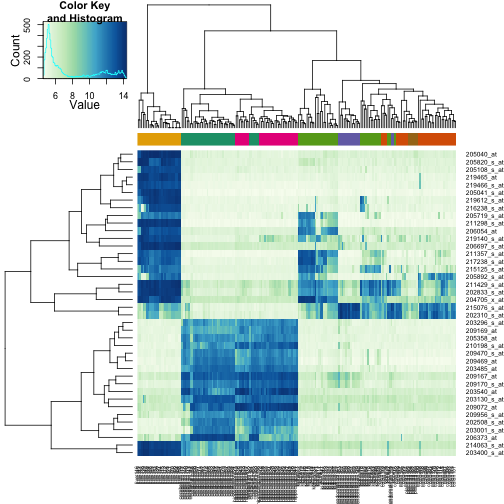
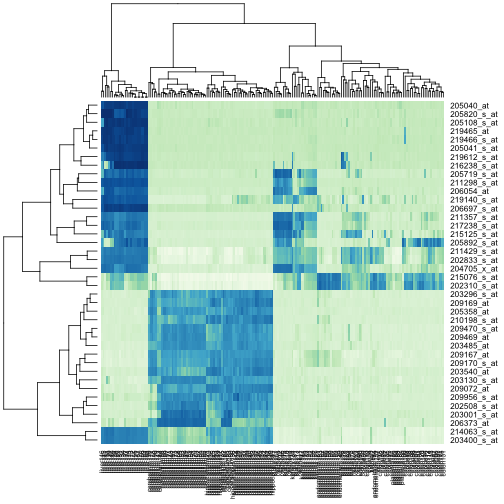
Heatmaps
Heatmaps are useful plots for visualizing the expression values for a
subset of genes over all the samples. The dendrogram on top and on
the side is a hierarchical clustering as we saw before. First we will
use the heatmap available in base R. First define a color palette.
# install.packages('RColorBrewer')
library(RColorBrewer)
hmcol <- colorRampPalette(brewer.pal(9, "GnBu"))(100)
Now, pick the genes with the top variance over all samples:
library(genefilter)
##
## Attaching package: 'genefilter'
##
## The following object is masked from 'package:base':
##
## anyNA
rv <- rowVars(exprs(t))
idx <- order(-rv)[1:40]
Now we can plot a heatmap of these genes:
heatmap(exprs(t)[idx, ], col = hmcol)
The heatmap.2 function in the gplots package on CRAN is a bit more
customizable, and stretches to fill the window. Here we add colors to
indicate the tissue on the top:
# install.packages('gplots')
library(gplots)
## KernSmooth 2.23 loaded
## Copyright M. P. Wand 1997-2009
##
## Attaching package: 'gplots'
##
## The following object is masked from 'package:stats':
##
## lowess
cols <- palette(brewer.pal(8, "Dark2"))[t$Tissue]
cbind(colnames(t), cols)
## cols
## [1,] "kidney1" "#66A61E"
## [2,] "kidney2" "#66A61E"
## [3,] "kidney3" "#66A61E"
## [4,] "kidney4" "#66A61E"
## [5,] "kidney5" "#66A61E"
## [6,] "kidney6" "#66A61E"
## [7,] "kidney7" "#66A61E"
## [8,] "kidney8" "#66A61E"
## [9,] "kidney9" "#66A61E"
## [10,] "kidney10" "#66A61E"
## [11,] "kidney11" "#66A61E"
## [12,] "kidney12" "#66A61E"
## [13,] "kidney13" "#66A61E"
## [14,] "kidney14" "#66A61E"
## [15,] "kidney15" "#66A61E"
## [16,] "kidney16" "#66A61E"
## [17,] "hippocampus17" "#E7298A"
## [18,] "hippocampus18" "#E7298A"
## [19,] "hippocampus19" "#E7298A"
## [20,] "hippocampus20" "#E7298A"
## [21,] "hippocampus21" "#E7298A"
## [22,] "hippocampus22" "#E7298A"
## [23,] "hippocampus23" "#E7298A"
## [24,] "hippocampus24" "#E7298A"
## [25,] "hippocampus25" "#E7298A"
## [26,] "hippocampus26" "#E7298A"
## [27,] "hippocampus27" "#E7298A"
## [28,] "hippocampus28" "#E7298A"
## [29,] "hippocampus29" "#E7298A"
## [30,] "hippocampus30" "#E7298A"
## [31,] "hippocampus31" "#E7298A"
## [32,] "hippocampus32" "#E7298A"
## [33,] "hippocampus33" "#E7298A"
## [34,] "hippocampus34" "#E7298A"
## [35,] "hippocampus35" "#E7298A"
## [36,] "hippocampus36" "#E7298A"
## [37,] "hippocampus37" "#E7298A"
## [38,] "hippocampus38" "#E7298A"
## [39,] "hippocampus39" "#E7298A"
## [40,] "hippocampus40" "#E7298A"
## [41,] "hippocampus41" "#E7298A"
## [42,] "hippocampus42" "#E7298A"
## [43,] "hippocampus43" "#E7298A"
## [44,] "hippocampus44" "#E7298A"
## [45,] "hippocampus45" "#E7298A"
## [46,] "hippocampus46" "#E7298A"
## [47,] "cerebellum47" "#1B9E77"
## [48,] "cerebellum48" "#1B9E77"
## [49,] "cerebellum49" "#1B9E77"
## [50,] "cerebellum50" "#1B9E77"
## [51,] "cerebellum51" "#1B9E77"
## [52,] "cerebellum52" "#1B9E77"
## [53,] "cerebellum53" "#1B9E77"
## [54,] "cerebellum54" "#1B9E77"
## [55,] "cerebellum55" "#1B9E77"
## [56,] "cerebellum56" "#1B9E77"
## [57,] "cerebellum57" "#1B9E77"
## [58,] "cerebellum58" "#1B9E77"
## [59,] "cerebellum59" "#1B9E77"
## [60,] "cerebellum60" "#1B9E77"
## [61,] "cerebellum61" "#1B9E77"
## [62,] "cerebellum62" "#1B9E77"
## [63,] "cerebellum63" "#1B9E77"
## [64,] "cerebellum64" "#1B9E77"
## [65,] "cerebellum65" "#1B9E77"
## [66,] "cerebellum66" "#1B9E77"
## [67,] "cerebellum67" "#1B9E77"
## [68,] "cerebellum68" "#1B9E77"
## [69,] "cerebellum69" "#1B9E77"
## [70,] "cerebellum70" "#1B9E77"
## [71,] "cerebellum71" "#1B9E77"
## [72,] "cerebellum72" "#1B9E77"
## [73,] "cerebellum73" "#1B9E77"
## [74,] "kidney74" "#66A61E"
## [75,] "kidney75" "#66A61E"
## [76,] "kidney76" "#66A61E"
## [77,] "kidney77" "#66A61E"
## [78,] "kidney78" "#66A61E"
## [79,] "kidney79" "#66A61E"
## [80,] "kidney80" "#66A61E"
## [81,] "kidney81" "#66A61E"
## [82,] "kidney82" "#66A61E"
## [83,] "kidney83" "#66A61E"
## [84,] "kidney84" "#66A61E"
## [85,] "kidney85" "#66A61E"
## [86,] "kidney86" "#66A61E"
## [87,] "colon87" "#D95F02"
## [88,] "colon88" "#D95F02"
## [89,] "colon89" "#D95F02"
## [90,] "colon90" "#D95F02"
## [91,] "colon91" "#D95F02"
## [92,] "colon92" "#D95F02"
## [93,] "colon93" "#D95F02"
## [94,] "colon94" "#D95F02"
## [95,] "colon95" "#D95F02"
## [96,] "colon96" "#D95F02"
## [97,] "colon97" "#D95F02"
## [98,] "colon98" "#D95F02"
## [99,] "colon99" "#D95F02"
## [100,] "colon100" "#D95F02"
## [101,] "colon101" "#D95F02"
## [102,] "colon102" "#D95F02"
## [103,] "colon103" "#D95F02"
## [104,] "colon104" "#D95F02"
## [105,] "colon105" "#D95F02"
## [106,] "colon106" "#D95F02"
## [107,] "colon107" "#D95F02"
## [108,] "colon108" "#D95F02"
## [109,] "colon109" "#D95F02"
## [110,] "colon110" "#D95F02"
## [111,] "colon111" "#D95F02"
## [112,] "colon112" "#D95F02"
## [113,] "colon113" "#D95F02"
## [114,] "colon114" "#D95F02"
## [115,] "colon115" "#D95F02"
## [116,] "colon116" "#D95F02"
## [117,] "colon117" "#D95F02"
## [118,] "colon118" "#D95F02"
## [119,] "colon119" "#D95F02"
## [120,] "colon120" "#D95F02"
## [121,] "kidney121" "#66A61E"
## [122,] "kidney122" "#66A61E"
## [123,] "kidney123" "#66A61E"
## [124,] "liver124" "#E6AB02"
## [125,] "liver125" "#E6AB02"
## [126,] "liver126" "#E6AB02"
## [127,] "kidney127" "#66A61E"
## [128,] "kidney128" "#66A61E"
## [129,] "kidney129" "#66A61E"
## [130,] "liver130" "#E6AB02"
## [131,] "liver131" "#E6AB02"
## [132,] "liver132" "#E6AB02"
## [133,] "kidney133" "#66A61E"
## [134,] "kidney134" "#66A61E"
## [135,] "cerebellum135" "#1B9E77"
## [136,] "hippocampus136" "#E7298A"
## [137,] "liver137" "#E6AB02"
## [138,] "liver138" "#E6AB02"
## [139,] "liver139" "#E6AB02"
## [140,] "liver140" "#E6AB02"
## [141,] "liver141" "#E6AB02"
## [142,] "liver142" "#E6AB02"
## [143,] "cerebellum143" "#1B9E77"
## [144,] "cerebellum144" "#1B9E77"
## [145,] "liver145" "#E6AB02"
## [146,] "liver146" "#E6AB02"
## [147,] "kidney147" "#66A61E"
## [148,] "kidney148" "#66A61E"
## [149,] "endometrium149" "#7570B3"
## [150,] "endometrium150" "#7570B3"
## [151,] "endometrium151" "#7570B3"
## [152,] "endometrium152" "#7570B3"
## [153,] "endometrium153" "#7570B3"
## [154,] "endometrium154" "#7570B3"
## [155,] "endometrium155" "#7570B3"
## [156,] "endometrium156" "#7570B3"
## [157,] "endometrium157" "#7570B3"
## [158,] "endometrium158" "#7570B3"
## [159,] "endometrium159" "#7570B3"
## [160,] "endometrium160" "#7570B3"
## [161,] "endometrium161" "#7570B3"
## [162,] "endometrium162" "#7570B3"
## [163,] "endometrium163" "#7570B3"
## [164,] "liver164" "#E6AB02"
## [165,] "liver165" "#E6AB02"
## [166,] "liver166" "#E6AB02"
## [167,] "liver167" "#E6AB02"
## [168,] "liver168" "#E6AB02"
## [169,] "liver169" "#E6AB02"
## [170,] "liver170" "#E6AB02"
## [171,] "liver171" "#E6AB02"
## [172,] "liver172" "#E6AB02"
## [173,] "liver173" "#E6AB02"
## [174,] "liver174" "#E6AB02"
## [175,] "liver175" "#E6AB02"
## [176,] "cerebellum176" "#1B9E77"
## [177,] "cerebellum177" "#1B9E77"
## [178,] "cerebellum178" "#1B9E77"
## [179,] "cerebellum179" "#1B9E77"
## [180,] "cerebellum180" "#1B9E77"
## [181,] "cerebellum181" "#1B9E77"
## [182,] "cerebellum182" "#1B9E77"
## [183,] "cerebellum183" "#1B9E77"
## [184,] "placenta184" "#A6761D"
## [185,] "placenta185" "#A6761D"
## [186,] "placenta186" "#A6761D"
## [187,] "placenta187" "#A6761D"
## [188,] "placenta188" "#A6761D"
## [189,] "placenta189" "#A6761D"
heatmap.2(exprs(t)[idx, ], trace = "none", ColSideColors = cols, col = hmcol)