Reproducible research: basic considerations
Overview
A key reason for contributing to open software and data for genome-scale is fostering reproducibility and extensibility of important findings. Two forms of reproducibility are
- concrete reproducibility: the published computational result can be exactly reproduced by independent investigators in possession of the code and data;
- amenability to replication: independent investigators equipped with the same experimental protocol, and comparable biological specimens and lab environment, will obtain qualitatively similar inferences.
There are many examples in modern genomic science where neither form of reproducibility is achieved, hence a burgeoning movement called “reproducible research” has had significant interaction with practitioners in genome biology.
The National Academy of Sciences has issued a report on statistical and methodological concerns in the domain of reproducibility:

A number of interesting infrastructure responses to reproducibility problems are noted by Victoria Stodden:

She also gives a treatment of statistical reproducibility:

Topics to be covered in this unit
We want to be able to foster reproducibility and extension of our work. To this end we will cover
- construction of R packages for software and annotation, that are versioned and can include formal tests for correctness of function operation
- use of knitr to create workflow documents – background in notes of Laurent Gatto
- use of the docker container discipline to specify and recover environments in which computations are performed
Additional commentary
The three themes underlying reproducibility research
1) providing code and data and environment to independent parties to diminish risk of analyses that are not reproducible
2) fortifying criteria of statistical soundness of analyses (study interpretations) to control risk of non-replicability of studies
3) doing 1) and 2) in ways that are cost-effective
Y. Benjamini, NAS workshop p. 47
[R]eproducibility is a property of a study, and replicability is a property of a result that can be proved only by inspecting other results of similar experiments. Therefore, the reproducibility of a result from a single study can be assured, and improving the statistical analysis can enhance its replicability.
Upshots:
- assuring reproducibility requires technique by the investigator
- enhancing replicability requires new approaches to statistical measurement of evidence
Additional considerations:
- Extensibility and transportability should not be divorced from reproducibility
- Reproducer/reader should be able to assess effects of modifications to queries and inferences
- This is a major motivation for Bioconductor’s commitments to platform-independence, continuous integration, and archiving of prior package versions
- Scalability should not be divorced from reproducbility
- Will a computation that took the author days or weeks to create be checked by independent parties?
- Does the work support a stepwise approach to verification?
- Results are reproducible in detail for a meaningful subproblem
- Results are reproducible in detail for a sequence of meaningful subproblems of increasing difficulty
- Computable documents (rmarkdown, jupyter, …) are important for pursuit of cost-effectiveness of reproducible research discipline
Boos: Replication probabilities are low for conventional thresholds
The left panel of this diagram illustrates a somewhat unexpected situation with statistical testing. The axis measures the replication probability for some simple experimental designs as a function of the -value obtained for an initial experiment. The solid line marked “1” traces the relationship between the -value and the replication probability for a one-sample test of the null hypothesis that the mean of a population following the normal distribution is zero. The sample size is 10, and the significance level is 5 percent. The figure shows that if the initial experiment produces a -value of 0.025, the probability that an identically designed experiment will achieve a -value no greater than 0.05 is about 70 percent. To achieve high probability of replication (say, greater than 90%), the initial experiment should have a -value less than 0.01.
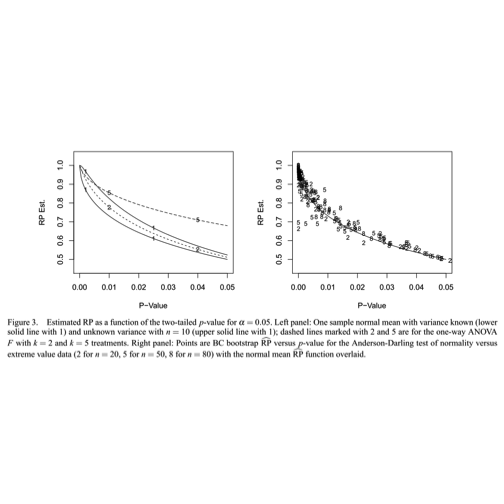
Boos and Stefanski (American Statistician, 65:4, 211) provide R code to trace the one-sample replication probability curve:
rp.t2 = function (pv, n, a = 0.05)
{
1 - pt(qt(1 - a/2, df = n - 1), df = n - 1, ncp = -qt(pv/2,
df = n - 1)) + pt(-qt(1 - a/2, df = n - 1), df = n -
1, ncp = -qt(pv/2, df = n - 1))
}
The upshot is that -values are statistically variable, but their uncertainty is not reckoned in conventional usage. Boos and Stefanski suggest that the order of magnitude of is fairly reliable but finer-grained distinctions are not.
Some material from the NAS workshop: Valen Johnson (PNAS paper)
In the following, UMPBT denotes ‘uniformly most powerful Bayesian test’. This is a relatively recent notion that has been described in an open publication by Valen Johnson Regardless of the reference to Bayesian concepts, the effects of pursuing more stringent criteria of significance must be addressed.
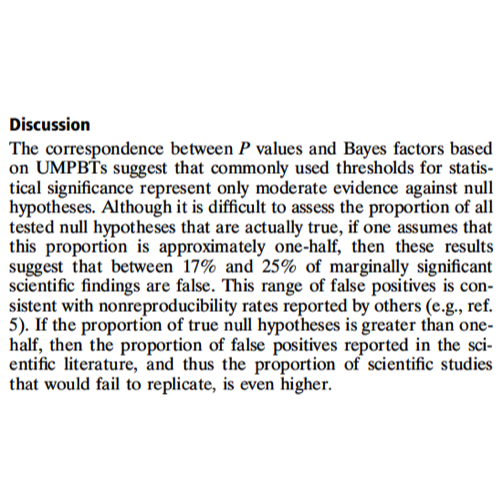
Costs of more stringent thresholds:

Upshots
- Statistical theory for enhancing replicability of analyses of independent experiments is substantial
- Costs of increasing replicability are non-trivial
- Preserving the reputation of advanced science may well justify the expenditure